Introduction to the Bass Diffusion Model#
What is the Bass Model?#
The Bass diffusion model, developed by Frank Bass in 1969, is a mathematical model that describes how new products get adopted in a population over time. It’s widely used in marketing to forecast sales of new products, especially when historical data is limited or non-existent.
The model captures the entire lifecycle of product adoption, from introduction to saturation, making it a powerful tool for product planning and marketing strategy development.
The Motivation Behind the Bass Model#
Before the Bass model, companies struggled to predict the adoption patterns of new products. Traditional forecasting methods often failed because they couldn’t account for the social dynamics that drive product adoption. Frank Bass recognized that product adoption follows a distinct pattern:
Initial slow growth: When a product first launches, adoption starts slowly
Rapid acceleration: As more people adopt, word-of-mouth spreads and adoption accelerates
Eventual saturation: Eventually, the market becomes saturated and adoption slows down
The Bass model provides a mathematical framework to capture these patterns, enabling businesses to make more informed decisions about production planning, inventory management, and marketing resource allocation.
Mathematical Formulation#
The Bass model is based on a differential equation that describes the rate of adoption:
Where:
\(F(t)\) is the installed base fraction (cumulative proportion of adopters)
\(f(t)\) is the rate of change of the installed base fraction (\(f(t) = F'(t)\))
\(p\) is the coefficient of innovation or external influence
\(q\) is the coefficient of imitation or internal influence
The solution to this equation gives the adoption curve:
The adoption rate at time \(t\) is given by:
Alternatively, this can be written as:
Key Components of the Bass Model Implementation#
The Bass model implementation in PyMC-Marketing consists of several key components:
Adopters - The number of new adoptions at time \(t\):
Innovators - Adoptions driven by external influence (advertising, etc.):
Imitators - Adoptions driven by internal influence (word-of-mouth):
Peak Adoption Time - When the adoption rate reaches its maximum:
The total number of adopters over time is the sum of innovators and imitators, which equals \(\text{adopters}(t)\). All of these components are directly implemented in the PyMC model, allowing us to analyze each aspect of the diffusion process separately.
Understanding the Relationship Between Components#
A key insight of the Bass model is how it decomposes adoption into two sources:
At each time point:
Innovators (\(m \cdot p \cdot (1 - F(t))\)) represents new adoptions coming from people who are influenced by external factors like advertising
Imitators (\(m \cdot q \cdot F(t) \cdot (1 - F(t))\)) represents new adoptions coming from people who are influenced by previous adopters
As time progresses:
Initially, innovators dominate the adoption process when few people have adopted (\(F(t)\) is small)
Later, imitators become the primary source of new adoptions as the word-of-mouth effect grows
Eventually, both decrease as the market approaches saturation (\(F(t)\) approaches 1)
The cumulative adoption at any time point is:
This means that as \(t \to \infty\), the cumulative adoption approaches the total market potential \(m\):
Therefore, the Bass model provides a complete accounting of the market:
At each time point, new adopters are either innovators or imitators
Over the entire product lifecycle, all potential adopters (m) eventually adopt the product
The model tracks both the adoption rate (new adopters per time period) and the cumulative adoption (total adopters to date)
This structure enables marketers to understand not just how many people will adopt over time, but also the driving forces behind adoption at different stages of the product lifecycle.
Understanding the Key Parameters#
The model has three main parameters:
Market potential (m): Total number of eventual adopters (the ultimate market size)
Innovation coefficient (p): Measures external influence like advertising and media - typically \(0.01-0.03\)
Imitation coefficient (q): Measures internal influence like word-of-mouth - typically \(0.3-0.5\)
Parameter Interpretation#
A higher p value indicates stronger external influence (advertising, marketing)
A higher q value indicates stronger internal influence (word-of-mouth, social interactions)
The ratio q/p indicates the relative strength of internal vs. external influences
The peak of adoption occurs at time
Innovators vs. Imitators#
The Bass model distinguishes between two types of adopters:
Innovators: People who adopt independently of others’ decisions, influenced mainly by mass media and external communications
Mathematically represented as: \(\text{innovators}(t) = m \cdot p \cdot (1 - F(p, q, t))\)
Imitators: People who adopt because of social influence and word-of-mouth from previous adopters
Mathematically represented as: \(\text{imitators}(t) = m \cdot q \cdot F(p, q, t) \cdot (1 - F(p, q, t))\)
Real-World Applications#
The Bass model has been successfully applied to forecast the adoption of various products and technologies:
Consumer durables: TVs, refrigerators, washing machines
Technology products: Smartphones, computers, software
Pharmaceutical products: New drugs and treatments
Entertainment products: Movies, games, streaming services
Services and subscriptions: Banking services, subscription plans
Business Value: Why the Bass Model Matters to Executives and Marketers#
From a business perspective, the Bass diffusion model provides substantial competitive advantages and ROI benefits:
1. Resource Optimization and Cash Flow Management#
Production Planning: Avoid costly overproduction or stockouts by accurately forecasting demand curves
Marketing Budget Allocation: Optimize spending across the product lifecycle, investing more during key inflection points
Supply Chain Efficiency: Coordinate with suppliers and distributors based on predicted adoption rates
Cash Flow Optimization: Better predict revenue streams, improving financial planning and investor relations
2. Strategic Decision Making#
Launch Timing: Determine the optimal time to enter a market based on diffusion patterns
Pricing Strategy: Implement dynamic pricing strategies aligned with the adoption curve
Competitive Analysis: Compare your product’s adoption parameters with competitors to identify strengths and weaknesses
Product Portfolio Management: Make informed decisions about when to phase out older products and introduce new ones
3. Risk Mitigation#
Scenario Planning: Test different market assumptions and external factors through parameter variations
Early Warning System: Identify deviations from expected adoption curves early, enabling faster intervention
Investment Justification: Provide data-driven forecasts to justify R&D and marketing investments to stakeholders
4. Performance Measurement#
Marketing Effectiveness: Measure the impact of marketing campaigns on the innovation coefficient (p)
Word-of-Mouth Strength: Quantify the power of your brand’s social influence through the imitation coefficient (q)
Total Market Potential: Validate or adjust your total addressable market estimates (m)
In today’s data-driven business environment, companies that effectively utilize models like Bass diffusion gain a significant competitive edge through more precise forecasting, better resource allocation, and strategic market timing.
Bayesian Extensions#
In this notebook, we show how to generate simulated data from the Bass model and fit a Bayesian model to it. The Bayesian formulation offers several advantages:
Uncertainty quantification through prior distributions on parameters
Hierarchical modeling for multiple products or markets
Incorporation of expert knowledge through informative priors
Full probability distributions for future adoption forecasts
What we’ll do in this notebook#
In this notebook, we’ll:
Set up parameters for a Bass model simulation
Generate simulated adoption data for multiple products
Fit the Bass model to our simulated data using PyMC
Visualize the adoption curves
Prepare Notebook#
from typing import Any
import arviz as az
import matplotlib.pyplot as plt
import numpy as np
import numpy.typing as npt
import pandas as pd
import pymc as pm
import xarray as xr
from pymc_marketing.bass.model import create_bass_model
from pymc_marketing.plot import plot_curve
from pymc_marketing.prior import Prior, Scaled
az.style.use("arviz-darkgrid")
plt.rcParams["figure.figsize"] = [12, 7]
plt.rcParams["figure.dpi"] = 100
%load_ext autoreload
%autoreload 2
%config InlineBackend.figure_format = "retina"
seed: int = sum(map(ord, "bass"))
rng: np.random.Generator = np.random.default_rng(seed=seed)
Setting Up Simulation Parameters#
First, we’ll set up the parameters for our simulation. This includes:
The time period for our simulation (in weeks)
The number of products to simulate
Start dates for the simulation period
def setup_simulation_parameters(
n_weeks: int = 52,
n_products: int = 9,
start_date: str = "2023-01-01",
cutoff_start_date: str = "2023-12-01",
) -> tuple[
npt.NDArray[np.int_],
pd.DatetimeIndex,
pd.DatetimeIndex,
list[str],
pd.Series,
dict[str, Any],
]:
"""Set up initial parameters for the Bass diffusion model simulation.
Parameters
----------
n_weeks : int
Number of weeks to simulate
n_products : int
Number of products to include in the simulation
start_date : str
Starting date for the simulation period
cutoff_start_date : str
Latest possible start date for products
Returns
-------
T : numpy.ndarray
Time array (weeks)
possible_dates : pandas.DatetimeIndex
All dates in the simulation period
possible_start_dates : pandas.DatetimeIndex
Possible start dates for products
products : list
List of product names
product_start : pandas.Series
Start date for each product
coords : dict
Coordinates for PyMC model
"""
# Set a seed for reproducibility
seed = sum(map(ord, "bass"))
rng = np.random.default_rng(seed)
# Create time array and date range
T = np.arange(n_weeks)
possible_dates = pd.date_range(start_date, freq="W-MON", periods=n_weeks)
cutoff_start_date = pd.to_datetime(cutoff_start_date)
cutoff_start_date = cutoff_start_date + pd.DateOffset(weeks=1)
possible_start_dates = possible_dates[possible_dates < cutoff_start_date]
# Generate product names and random start dates
products = [f"P{i}" for i in range(n_products)]
product_start = pd.Series(
rng.choice(possible_start_dates, size=len(products)),
index=pd.Index(products, name="product"),
)
coords = {"T": T, "product": products}
return T, possible_dates, possible_start_dates, products, product_start, coords
Creating Prior Distributions#
For our Bayesian Bass model, we need to specify prior distributions for the key parameters:
m (market potential): How many units can potentially be sold in total
p (innovation coefficient): Rate of adoption from external influences
q (imitation coefficient): Rate of adoption from internal/social influences
likelihood: The probability distribution that models the observed adoption data
For the market potential m we use a scaling trick to specify a scale-free prior and then add a global factor:
def create_bass_priors(factor: float) -> dict[str, Prior | Scaled]:
"""Define prior distributions for the Bass model parameters.
Returns
-------
dict
Dictionary of prior distributions for m, p, q, and likelihood
Notes
-----
- m: Market potential (scaled Gamma distribution)
- p: Innovation coefficient (Beta distribution)
- q: Imitation coefficient (Beta distribution)
- likelihood: Observation model (Negative Binomial)
"""
return {
# We use a scaled Gamma distribution for the market potential.
"m": Scaled(Prior("Gamma", mu=1, sigma=0.1, dims="product"), factor=factor),
"p": Prior("Beta", mu=0.03, dims="product").constrain(lower=0.01, upper=0.03),
"q": Prior("Beta", dims="product").constrain(lower=0.3, upper=0.5),
"likelihood": Prior("NegativeBinomial", n=1.5, dims="product"),
}
Let’s generate and visualize the priors.
FACTOR = 50_000
priors = create_bass_priors(factor=FACTOR)
/Users/will/mamba/envs/pymc-marketing-dev/lib/python3.10/site-packages/preliz/distributions/beta.py:127: RuntimeWarning: invalid value encountered in scalar divide
mu = alpha / alpha_plus_beta
/Users/will/mamba/envs/pymc-marketing-dev/lib/python3.10/site-packages/preliz/distributions/beta.py:128: RuntimeWarning: invalid value encountered in scalar divide
sigma = (alpha * beta) ** 0.5 / alpha_plus_beta / (alpha_plus_beta + 1) ** 0.5
The requested mass is 0.95, but the computed one is 0.528
fig, ax = plt.subplots(nrows=2, ncols=1, figsize=(15, 12))
priors["p"].preliz.plot_pdf(ax=ax[0])
ax[0].set(title="Innovation Coefficient (p)")
priors["q"].preliz.plot_pdf(ax=ax[1])
ax[1].set(title="Imitation Coefficient (q)")
fig.suptitle(
"Prior Distributions for Bass Model Parameters",
fontsize=18,
fontweight="bold",
y=0.95,
);
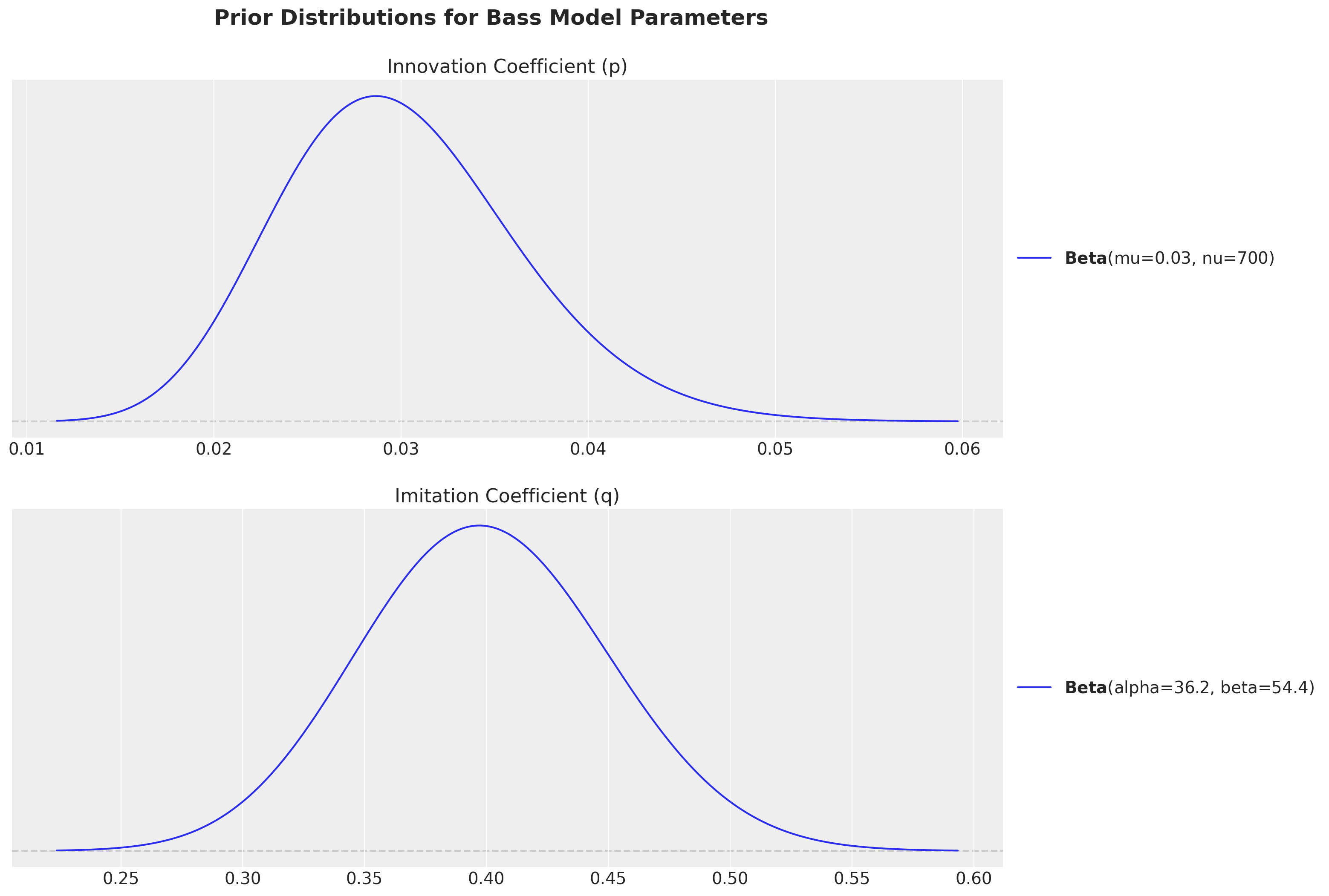
Observe we have chosen the priors within the usual ranges of empirical studies:
Innovation coefficient (p): Measures external influence like advertising and media - typically \(0.01-0.03\)
Imitation coefficient (q): Measures internal influence like word-of-mouth - typically \(0.3-0.5\)
Generate Synthetic Data#
With the generative Bass model, we can generate a synthetic dataset by sampling from the prior and choosing one particular sample to use as observed data. For this purpose we define two auxiliary functions.
def sample_prior_bass_data(model: pm.Model) -> xr.DataArray:
"""Generate a sample from the prior predictive distribution of the Bass model.
Parameters
----------
model : pymc.Model
The PyMC model to sample from
Returns
-------
xarray.DataArray
Simulated adoption data
"""
with model:
idata = pm.sample_prior_predictive(random_seed=rng)
return idata["prior"]["y"].sel(chain=0, draw=0)
def transform_to_actual_dates(bass_data, product_start, possible_dates) -> pd.DataFrame:
"""Transform simulation data from time index to calendar dates.
Parameters
----------
bass_data : xarray.DataArray
Simulated bass model data
product_start : pandas.Series
Start date for each product
possible_dates : pandas.DatetimeIndex
All dates in the simulation period
Returns
-------
pandas.DataFrame
Adoption data with actual calendar dates
"""
bass_data = bass_data.to_dataset()
bass_data["product_start"] = product_start.to_xarray()
df_bass_data = (
bass_data.to_dataframe().drop(columns=["chain", "draw"]).reset_index()
)
df_bass_data["actual_date"] = df_bass_data["product_start"] + pd.to_timedelta(
7 * df_bass_data["T"], unit="days"
)
return (
df_bass_data.set_index(["actual_date", "product"])
.y.unstack(fill_value=0)
.reindex(possible_dates, fill_value=0)
)
Now we can generate the observed data:
# Setup simulation parameters
T, possible_dates, _, products, product_start, coords = setup_simulation_parameters()
# Create and configure the Bass model
generative_model = create_bass_model(t=T, coords=coords, observed=None, priors=priors)
# Sample and select one "observed" dataset.
bass_data = sample_prior_bass_data(generative_model)
actual_data = transform_to_actual_dates(bass_data, product_start, possible_dates)
Sampling: [m_unscaled, p, q, y]
The actual_data data frame has the typical format of a real dataset.
actual_data
| product | P0 | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 |
|---|---|---|---|---|---|---|---|---|---|
| 2023-01-02 | 0 | 0 | 451 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2023-01-09 | 0 | 0 | 56 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2023-01-16 | 0 | 0 | 1948 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2023-01-23 | 0 | 0 | 3742 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2023-01-30 | 0 | 239 | 156 | 0 | 0 | 137 | 0 | 0 | 0 |
| 2023-02-06 | 0 | 4199 | 4732 | 0 | 0 | 2208 | 0 | 0 | 0 |
| 2023-02-13 | 0 | 1646 | 6477 | 0 | 0 | 1671 | 0 | 0 | 0 |
| 2023-02-20 | 0 | 1810 | 2824 | 0 | 0 | 780 | 0 | 0 | 0 |
| 2023-02-27 | 0 | 17004 | 2583 | 1485 | 0 | 6002 | 0 | 0 | 0 |
| 2023-03-06 | 0 | 4956 | 2508 | 2679 | 0 | 8211 | 0 | 0 | 0 |
| 2023-03-13 | 1277 | 7956 | 3255 | 2340 | 0 | 2404 | 0 | 0 | 0 |
| 2023-03-20 | 1403 | 4926 | 1030 | 3838 | 0 | 3646 | 0 | 0 | 0 |
| 2023-03-27 | 1818 | 1091 | 3145 | 1443 | 0 | 13557 | 0 | 0 | 0 |
| 2023-04-03 | 2865 | 4696 | 613 | 85 | 0 | 2857 | 0 | 0 | 0 |
| 2023-04-10 | 1410 | 8236 | 717 | 2858 | 0 | 3112 | 0 | 0 | 0 |
| 2023-04-17 | 1472 | 499 | 77 | 3519 | 0 | 3920 | 0 | 0 | 0 |
| 2023-04-24 | 11868 | 1783 | 340 | 2431 | 0 | 1661 | 0 | 0 | 0 |
| 2023-05-01 | 4669 | 102 | 314 | 8487 | 0 | 2908 | 0 | 0 | 0 |
| 2023-05-08 | 3423 | 61 | 210 | 1479 | 0 | 2729 | 0 | 0 | 0 |
| 2023-05-15 | 3625 | 1226 | 11 | 1544 | 0 | 2125 | 0 | 0 | 0 |
| 2023-05-22 | 1591 | 256 | 110 | 391 | 0 | 1164 | 0 | 0 | 0 |
| 2023-05-29 | 3351 | 149 | 78 | 2397 | 0 | 849 | 0 | 0 | 0 |
| 2023-06-05 | 1246 | 205 | 9 | 205 | 0 | 1060 | 0 | 0 | 0 |
| 2023-06-12 | 1805 | 106 | 22 | 187 | 0 | 229 | 0 | 0 | 0 |
| 2023-06-19 | 1353 | 75 | 15 | 131 | 0 | 38 | 0 | 0 | 0 |
| 2023-06-26 | 457 | 48 | 5 | 116 | 0 | 88 | 0 | 0 | 0 |
| 2023-07-03 | 170 | 46 | 8 | 108 | 0 | 79 | 0 | 0 | 0 |
| 2023-07-10 | 44 | 67 | 6 | 57 | 0 | 16 | 0 | 0 | 0 |
| 2023-07-17 | 271 | 8 | 1 | 37 | 0 | 14 | 0 | 0 | 0 |
| 2023-07-24 | 42 | 0 | 1 | 4 | 0 | 19 | 0 | 0 | 2578 |
| 2023-07-31 | 32 | 6 | 1 | 9 | 0 | 0 | 0 | 0 | 1418 |
| 2023-08-07 | 93 | 5 | 0 | 3 | 0 | 10 | 0 | 0 | 1005 |
| 2023-08-14 | 83 | 1 | 1 | 0 | 0 | 4 | 0 | 0 | 2560 |
| 2023-08-21 | 47 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 3666 |
| 2023-08-28 | 8 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 13816 |
| 2023-09-04 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 2496 | 7358 |
| 2023-09-11 | 4 | 0 | 0 | 0 | 0 | 0 | 556 | 1266 | 6785 |
| 2023-09-18 | 2 | 0 | 0 | 0 | 0 | 3 | 2151 | 3637 | 2370 |
| 2023-09-25 | 2 | 0 | 0 | 0 | 0 | 0 | 4980 | 1431 | 1351 |
| 2023-10-02 | 0 | 1 | 0 | 1 | 0 | 0 | 2124 | 507 | 2038 |
| 2023-10-09 | 0 | 0 | 0 | 0 | 0 | 0 | 14239 | 1632 | 3441 |
| 2023-10-16 | 0 | 0 | 0 | 0 | 0 | 0 | 25224 | 4681 | 562 |
| 2023-10-23 | 0 | 0 | 0 | 0 | 0 | 0 | 17233 | 2095 | 3801 |
| 2023-10-30 | 0 | 0 | 0 | 0 | 1042 | 0 | 4728 | 1408 | 446 |
| 2023-11-06 | 0 | 0 | 0 | 0 | 1776 | 0 | 1890 | 1890 | 487 |
| 2023-11-13 | 0 | 0 | 0 | 0 | 1697 | 0 | 1058 | 3665 | 673 |
| 2023-11-20 | 0 | 0 | 0 | 0 | 6648 | 0 | 943 | 391 | 169 |
| 2023-11-27 | 0 | 0 | 0 | 0 | 4746 | 0 | 605 | 2720 | 374 |
| 2023-12-04 | 0 | 0 | 0 | 0 | 1843 | 0 | 1554 | 1382 | 98 |
| 2023-12-11 | 0 | 0 | 0 | 0 | 3598 | 0 | 2304 | 147 | 151 |
| 2023-12-18 | 0 | 0 | 0 | 0 | 6576 | 0 | 124 | 2582 | 42 |
| 2023-12-25 | 0 | 0 | 0 | 0 | 3610 | 0 | 215 | 110 | 160 |
On the other hand, the bass_data has the same data as arrays indexed by time (relative) and product.
Let’s visualize both.
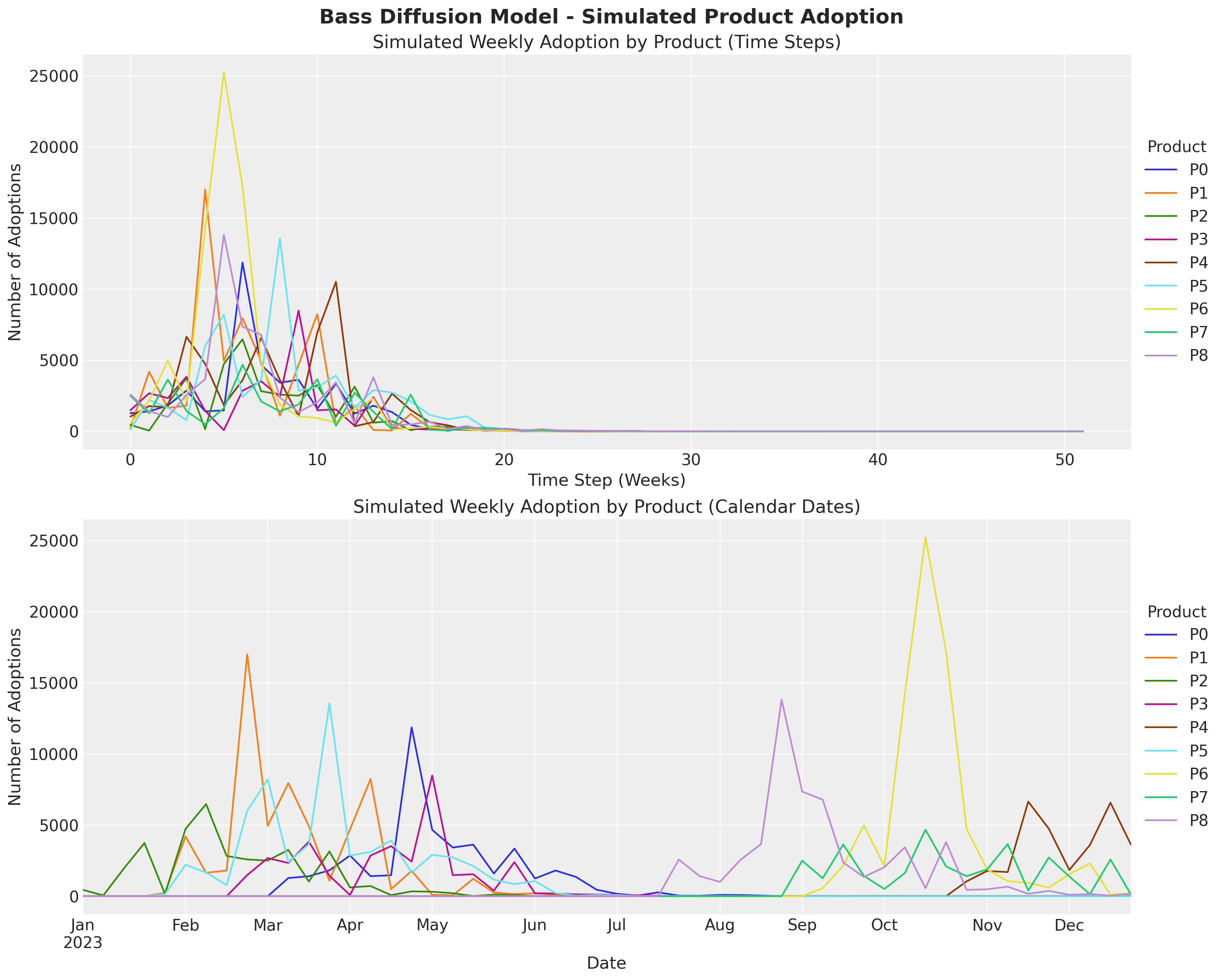
fig, ax = plt.subplots(
nrows=2, ncols=1, figsize=(15, 12), sharex=False, sharey=True, layout="constrained"
)
# Plot raw simulated data (by time step)
bass_data.to_series().unstack().plot(ax=ax[0])
ax[0].legend(
title="Product", title_fontsize=14, loc="center left", bbox_to_anchor=(1, 0.5)
)
ax[0].set(
title="Simulated Weekly Adoption by Product (Time Steps)",
xlabel="Time Step (Weeks)",
ylabel="Number of Adoptions",
)
# Plot data with actual calendar dates
actual_data.plot(ax=ax[1])
ax[1].legend(
title="Product", title_fontsize=14, loc="center left", bbox_to_anchor=(1, 0.5)
)
ax[1].set(
title="Simulated Weekly Adoption by Product (Calendar Dates)",
xlabel="Date",
ylabel="Number of Adoptions",
)
fig.suptitle(
"Bass Diffusion Model - Simulated Product Adoption", fontsize=18, fontweight="bold"
);
Fit the Model#
We are now ready to fit the model and generate the posterior predictive distributions.
# We condition the model on observed data.
with pm.observe(generative_model, {"y": bass_data.values}) as model:
idata = pm.sample(
tune=1_500,
draws=2_000,
chains=4,
nuts_sampler="nutpie",
compile_kwargs={"mode": "NUMBA"},
random_seed=rng,
)
idata.extend(
pm.sample_posterior_predictive(
idata, model=model, extend_inferencedata=True, random_seed=rng
)
)
Sampling ... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 / 0:00:01
We do not have any divergences. Let’s look at the summary of the parameters.
az.summary(data=idata, var_names=["p", "q", "m"])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| p[P0] | 0.026 | 0.005 | 0.018 | 0.036 | 0.000 | 0.000 | 10750.0 | 5899.0 | 1.0 |
| p[P1] | 0.030 | 0.005 | 0.020 | 0.039 | 0.000 | 0.000 | 11579.0 | 6089.0 | 1.0 |
| p[P2] | 0.027 | 0.005 | 0.017 | 0.036 | 0.000 | 0.000 | 11292.0 | 5970.0 | 1.0 |
| p[P3] | 0.030 | 0.005 | 0.020 | 0.040 | 0.000 | 0.000 | 12126.0 | 6250.0 | 1.0 |
| p[P4] | 0.026 | 0.005 | 0.018 | 0.035 | 0.000 | 0.000 | 12027.0 | 6287.0 | 1.0 |
| p[P5] | 0.021 | 0.004 | 0.013 | 0.028 | 0.000 | 0.000 | 10939.0 | 5817.0 | 1.0 |
| p[P6] | 0.032 | 0.005 | 0.022 | 0.042 | 0.000 | 0.000 | 11071.0 | 5834.0 | 1.0 |
| p[P7] | 0.030 | 0.005 | 0.020 | 0.039 | 0.000 | 0.000 | 12427.0 | 5130.0 | 1.0 |
| p[P8] | 0.030 | 0.005 | 0.021 | 0.040 | 0.000 | 0.000 | 11252.0 | 6353.0 | 1.0 |
| q[P0] | 0.402 | 0.019 | 0.368 | 0.439 | 0.000 | 0.000 | 10124.0 | 5923.0 | 1.0 |
| q[P1] | 0.394 | 0.019 | 0.360 | 0.429 | 0.000 | 0.000 | 10431.0 | 6172.0 | 1.0 |
| q[P2] | 0.397 | 0.019 | 0.363 | 0.434 | 0.000 | 0.000 | 11818.0 | 6179.0 | 1.0 |
| q[P3] | 0.450 | 0.022 | 0.413 | 0.493 | 0.000 | 0.000 | 12285.0 | 6374.0 | 1.0 |
| q[P4] | 0.354 | 0.017 | 0.324 | 0.386 | 0.000 | 0.000 | 11480.0 | 6068.0 | 1.0 |
| q[P5] | 0.385 | 0.018 | 0.350 | 0.418 | 0.000 | 0.000 | 9830.0 | 5566.0 | 1.0 |
| q[P6] | 0.430 | 0.020 | 0.393 | 0.468 | 0.000 | 0.000 | 12521.0 | 6256.0 | 1.0 |
| q[P7] | 0.344 | 0.017 | 0.311 | 0.375 | 0.000 | 0.000 | 12231.0 | 5460.0 | 1.0 |
| q[P8] | 0.339 | 0.017 | 0.307 | 0.369 | 0.000 | 0.000 | 10948.0 | 6146.0 | 1.0 |
| m[P0] | 49568.996 | 4491.414 | 40760.951 | 57490.821 | 41.520 | 55.743 | 11720.0 | 5933.0 | 1.0 |
| m[P1] | 51816.841 | 4487.349 | 43766.599 | 60653.332 | 41.988 | 55.334 | 11242.0 | 5793.0 | 1.0 |
| m[P2] | 47118.829 | 4436.392 | 39310.947 | 55843.304 | 41.551 | 54.987 | 11186.0 | 5118.0 | 1.0 |
| m[P3] | 48429.471 | 4518.438 | 40231.737 | 57157.439 | 41.948 | 54.340 | 11472.0 | 5854.0 | 1.0 |
| m[P4] | 51994.167 | 4501.491 | 43959.011 | 60680.809 | 43.404 | 54.416 | 10713.0 | 5973.0 | 1.0 |
| m[P5] | 53276.493 | 4568.591 | 45196.257 | 62426.608 | 46.587 | 61.073 | 9433.0 | 5492.0 | 1.0 |
| m[P6] | 53087.905 | 4612.168 | 44525.164 | 61948.611 | 45.914 | 58.424 | 10125.0 | 5873.0 | 1.0 |
| m[P7] | 47684.913 | 4315.345 | 39879.468 | 55899.426 | 40.015 | 50.408 | 11727.0 | 6210.0 | 1.0 |
| m[P8] | 51232.596 | 4464.032 | 42849.916 | 59268.568 | 43.671 | 57.224 | 10498.0 | 5213.0 | 1.0 |
_ = az.plot_trace(
data=idata,
var_names=["p", "q", "m"],
compact=True,
backend_kwargs={"figsize": (12, 7), "layout": "constrained"},
)
plt.gcf().suptitle("Model Trace", fontsize=16);
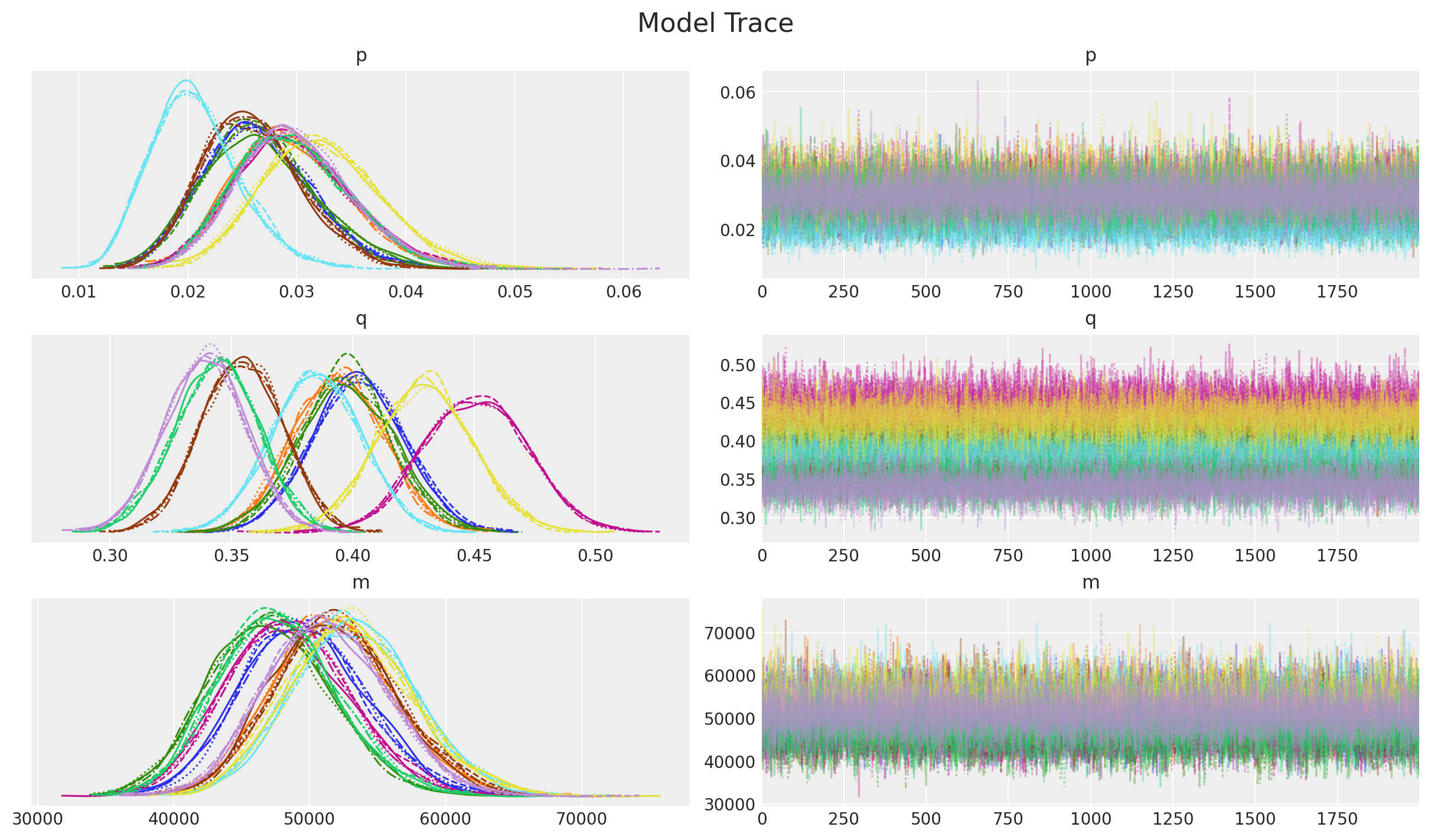
Overall, the diagnostics and trace look good.
Next, we look into the posterior distributions of the parameters.
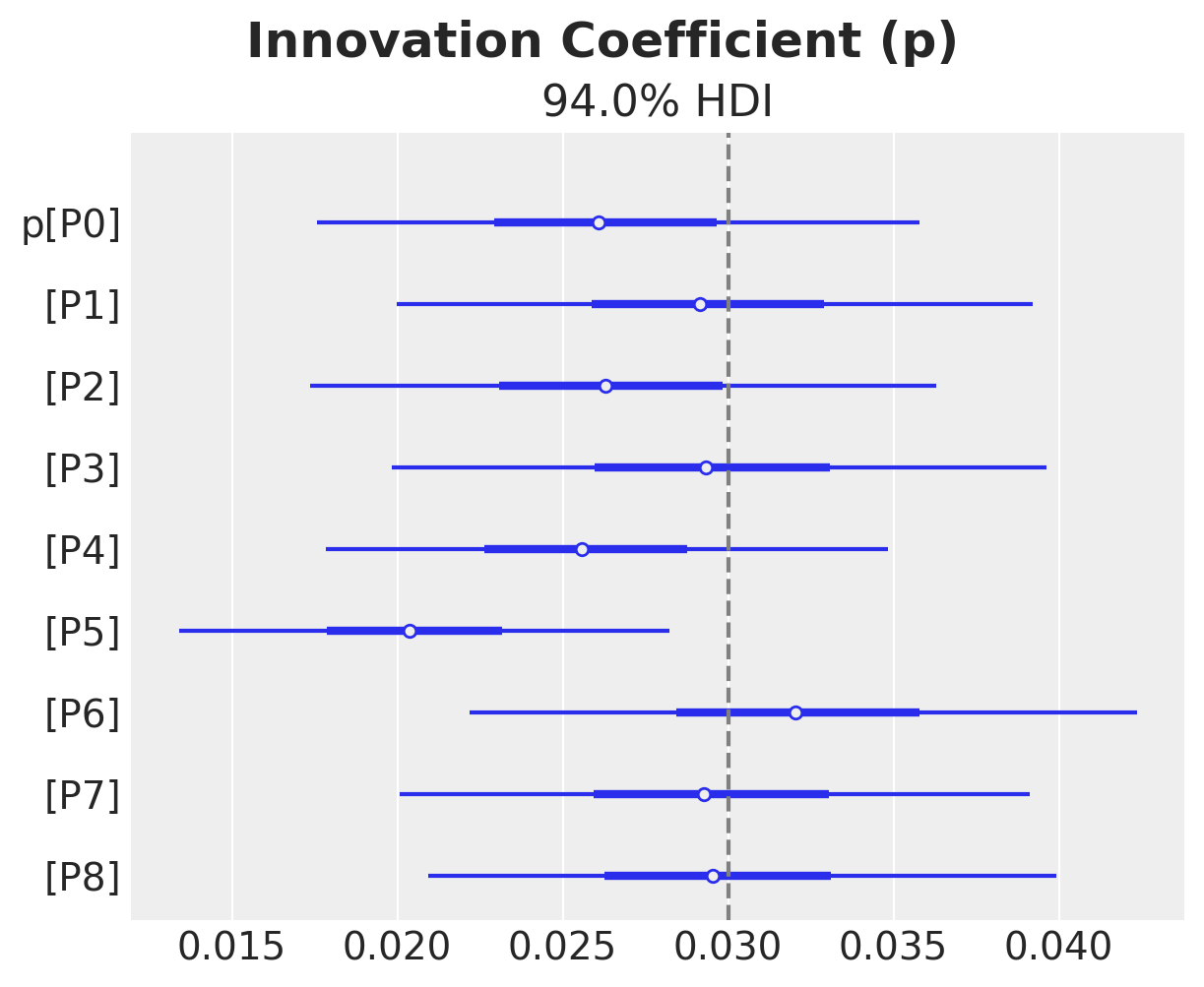
ax, *_ = az.plot_forest(idata["posterior"]["p"], combined=True)
ax.axvline(x=priors["p"].parameters["mu"], color="gray", linestyle="--")
ax.get_figure().suptitle("Innovation Coefficient (p)", fontsize=18, fontweight="bold")
Text(0.5, 0.98, 'Innovation Coefficient (p)')
ax, *_ = az.plot_forest(idata["posterior"]["q"], combined=True)
ax.axvline(x=priors["q"].preliz.mean(), color="gray", linestyle="--")
ax.get_figure().suptitle("Imitation Coefficient (q)", fontsize=18, fontweight="bold")
Text(0.5, 0.98, 'Imitation Coefficient (q)')
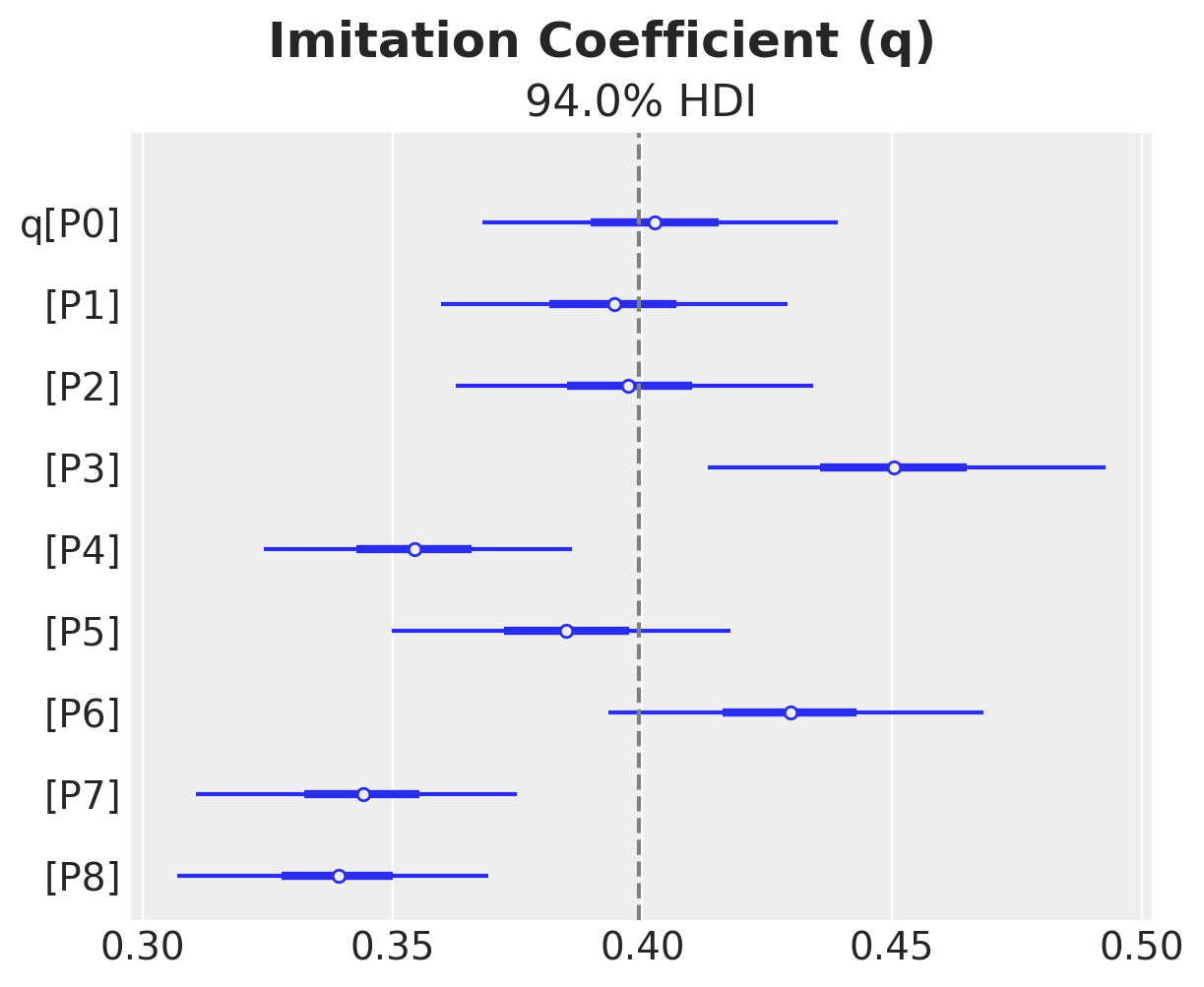
We do see some heterogeneity in the parameters, but overall they are centered around the true values (from the generative model).
Examining Posterior Predictions for Specific Products#
Let’s look at the posterior predictive distributions to see how well our model captures the simulated data.
fig, axes = plt.subplots(
nrows=3, ncols=3, figsize=(15, 12), sharex=True, sharey=True, layout="constrained"
)
idata["posterior_predictive"]["y"].pipe(plot_curve, {"T"}, axes=axes)
for i, ax in enumerate(axes.flatten()):
ax.plot(T, bass_data[:, i], color="black")
fig.suptitle("Posterior Predictive vs Observed Data", fontsize=18, fontweight="bold");
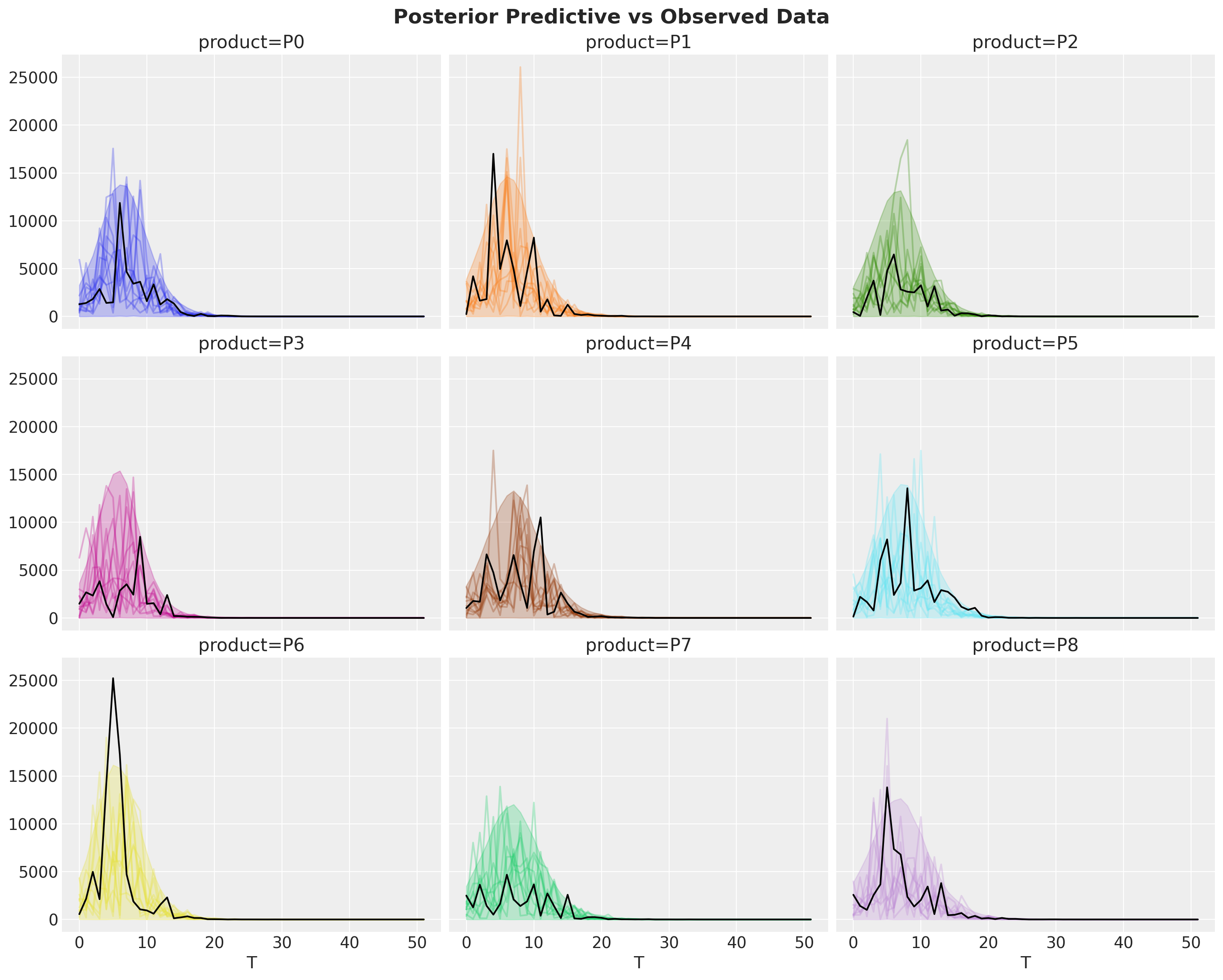
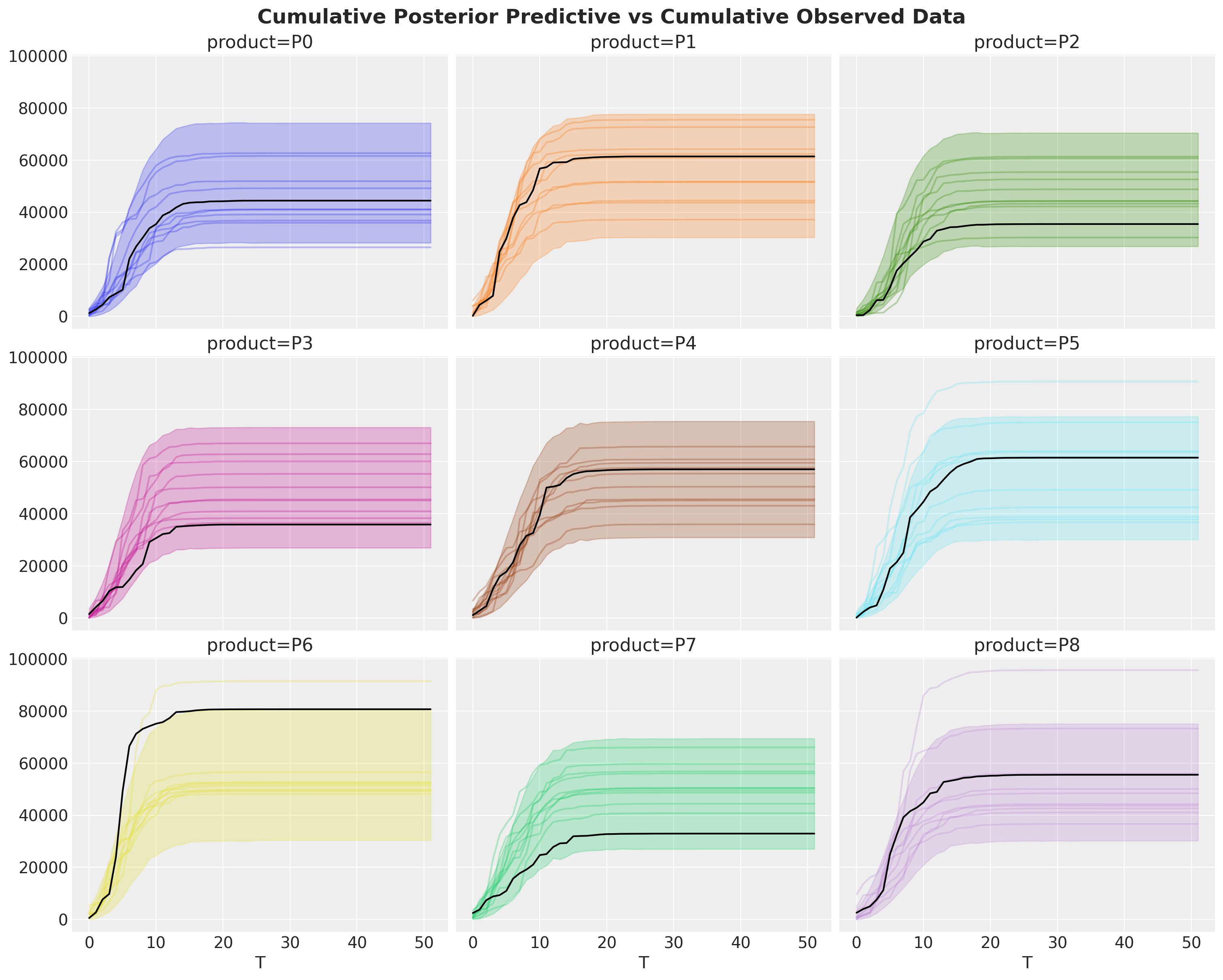
fig, axes = plt.subplots(
nrows=3, ncols=3, figsize=(15, 12), sharex=True, sharey=True, layout="constrained"
)
idata["posterior_predictive"]["y"].cumsum(dim="T").pipe(plot_curve, {"T"}, axes=axes)
for i, ax in enumerate(axes.flatten()):
ax.plot(T, bass_data[:, i].cumsum(), color="black")
fig.suptitle(
"Cumulative Posterior Predictive vs Cumulative Observed Data",
fontsize=18,
fontweight="bold",
);
observed_cumulative = bass_data.cumsum(dim="T").isel(T=-1).to_series()
ref_val = {
"m": [
{"product": name, "ref_val": value}
for name, value in observed_cumulative.items()
]
}
az.plot_posterior(
idata.posterior,
var_names=["m"],
backend_kwargs=dict(sharex=True, layout="constrained", figsize=(15, 12)),
ref_val=ref_val,
)
max_T = bass_data.coords["T"].max().item()
fig = plt.gcf()
fig.suptitle(
f"Estimated Market Cap (m) vs Observed Cumulative at T={max_T}",
fontsize=18,
fontweight="bold",
);
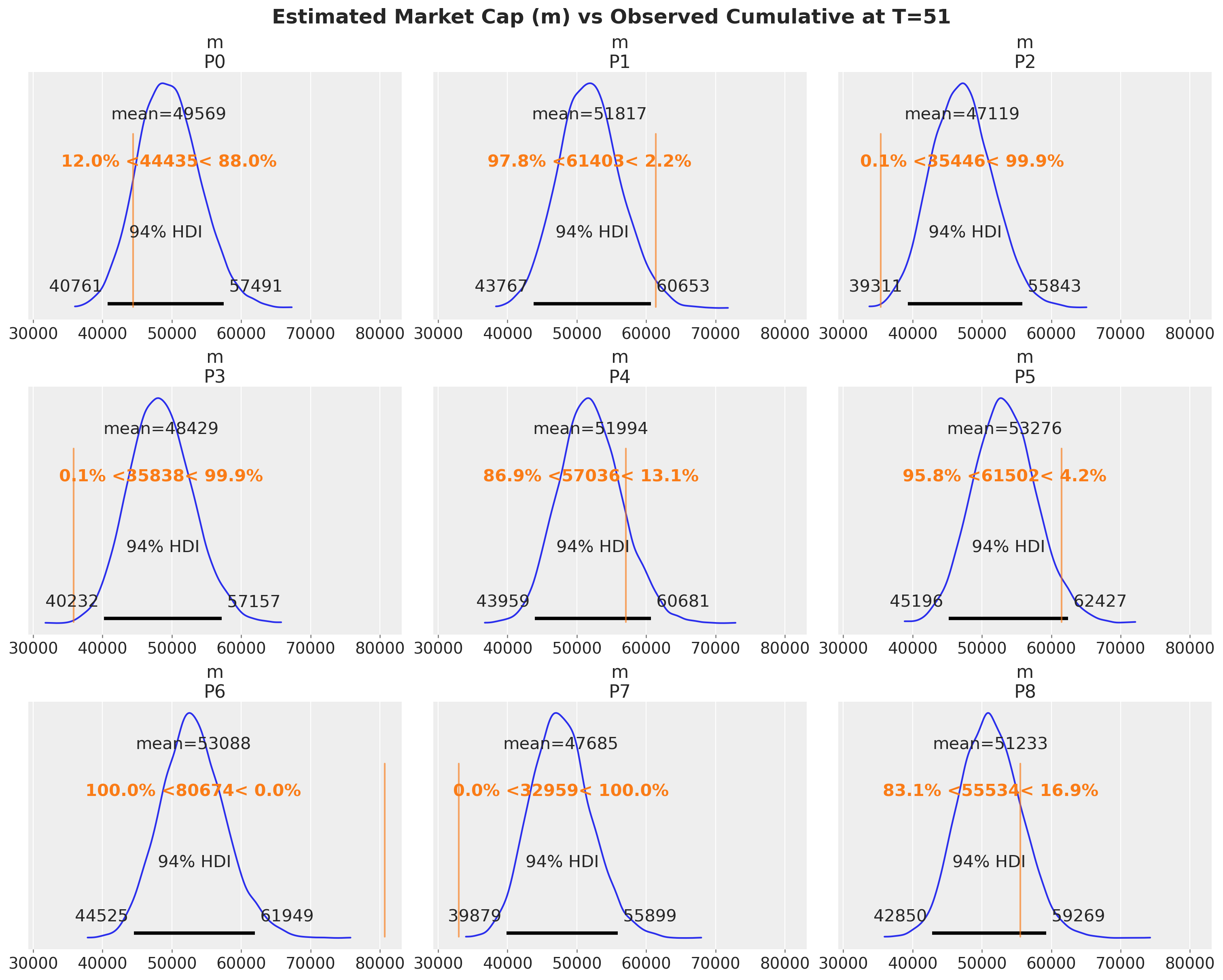
Overall, the model does a good job of capturing the data.
Next, we look into the adopters, which represent the expected value of the likelihood.
fig, axes = plt.subplots(
nrows=3, ncols=3, figsize=(15, 12), sharex=True, sharey=True, layout="constrained"
)
idata["posterior"]["adopters"].pipe(plot_curve, {"T"}, axes=axes)
for i, ax in enumerate(axes.flatten()):
ax.plot(T, bass_data[:, i], color="black")
fig.suptitle("Adopters vs Observed Data", fontsize=18, fontweight="bold");
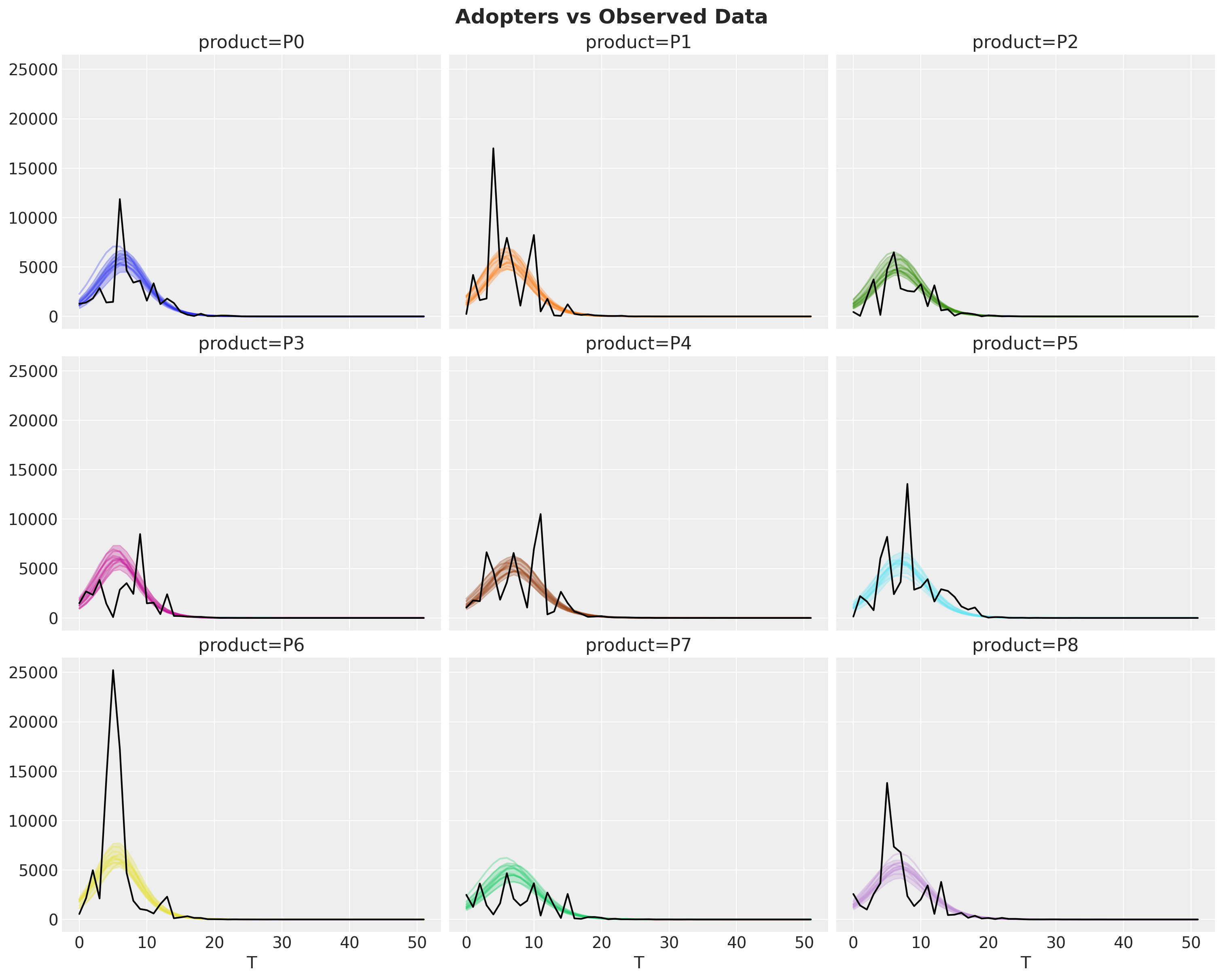
This show the fit is indeed quite reasonable.
We can also evaluate the model goodness by looking into the cumulative data:
Note
Remember that the adopters is the mean of the distribution so we see some cumulative curves above and some below.
Look at the idata["posterior_predictive"]["y"] for the observed data.
fig, axes = plt.subplots(
nrows=3, ncols=3, figsize=(15, 12), sharex=True, sharey=True, layout="constrained"
)
idata["posterior"]["adopters"].cumsum(dim="T").pipe(plot_curve, {"T"}, axes=axes)
for i, ax in enumerate(axes.flatten()):
ax.plot(T, bass_data[:, i].cumsum(), color="black")
fig.suptitle("Adopters Cumulative vs Observed Data", fontsize=18, fontweight="bold");
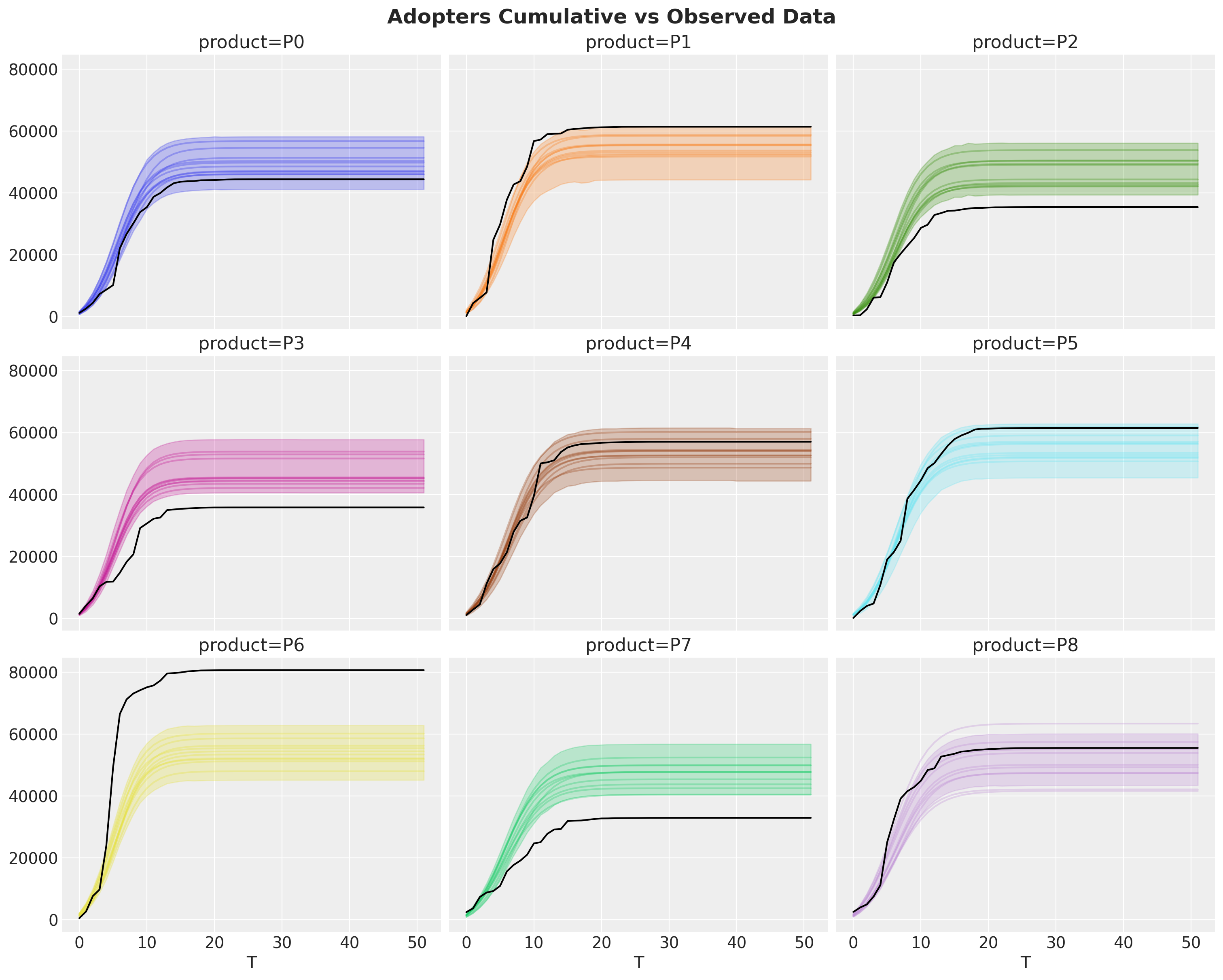
We can enhance this view by looking into the components of the model: innovators and imitators (in orange and green, respectively).
fig, axes = plt.subplots(
nrows=3, ncols=3, figsize=(15, 12), sharex=True, sharey=True, layout="constrained"
)
idata["posterior"]["adopters"].cumsum(dim="T").pipe(
plot_curve, {"T"}, colors=3 * 3 * ["C0"], axes=axes
)
idata["posterior"]["innovators"].pipe(
plot_curve, {"T"}, colors=3 * 3 * ["C1"], axes=axes
)
idata["posterior"]["imitators"].pipe(
plot_curve, {"T"}, colors=3 * 3 * ["C2"], axes=axes
)
for i, ax in enumerate(axes.flatten()):
ax.plot(T, bass_data[:, i].cumsum(), color="black")
fig.suptitle("Innovators vs Imitators", fontsize=18, fontweight="bold");
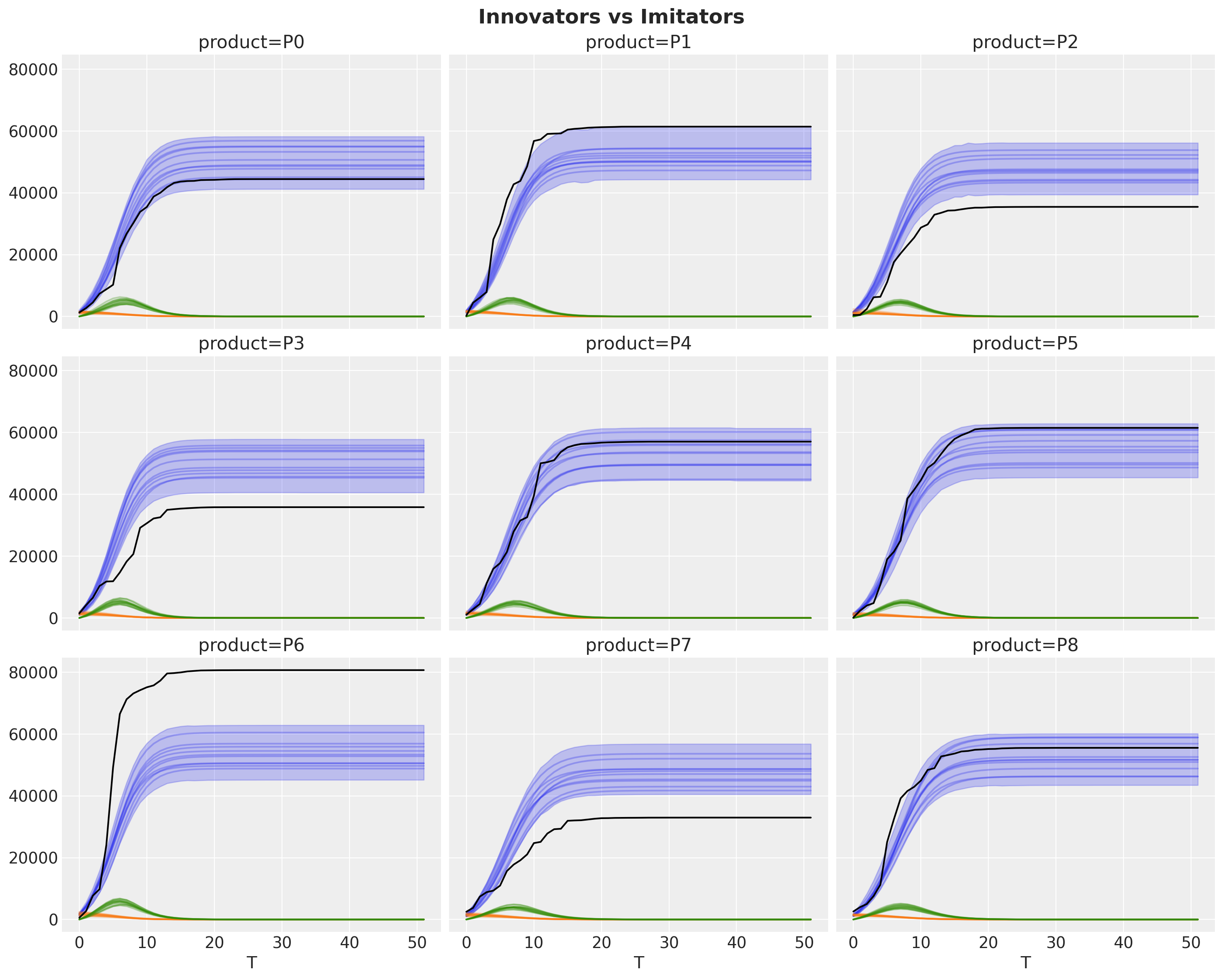
Finally, we can inspect the peak of the adoption curve.
ax, *_ = az.plot_forest(idata["posterior"]["peak"], combined=True)
ax.get_figure().suptitle("Peak", fontsize=18, fontweight="bold");
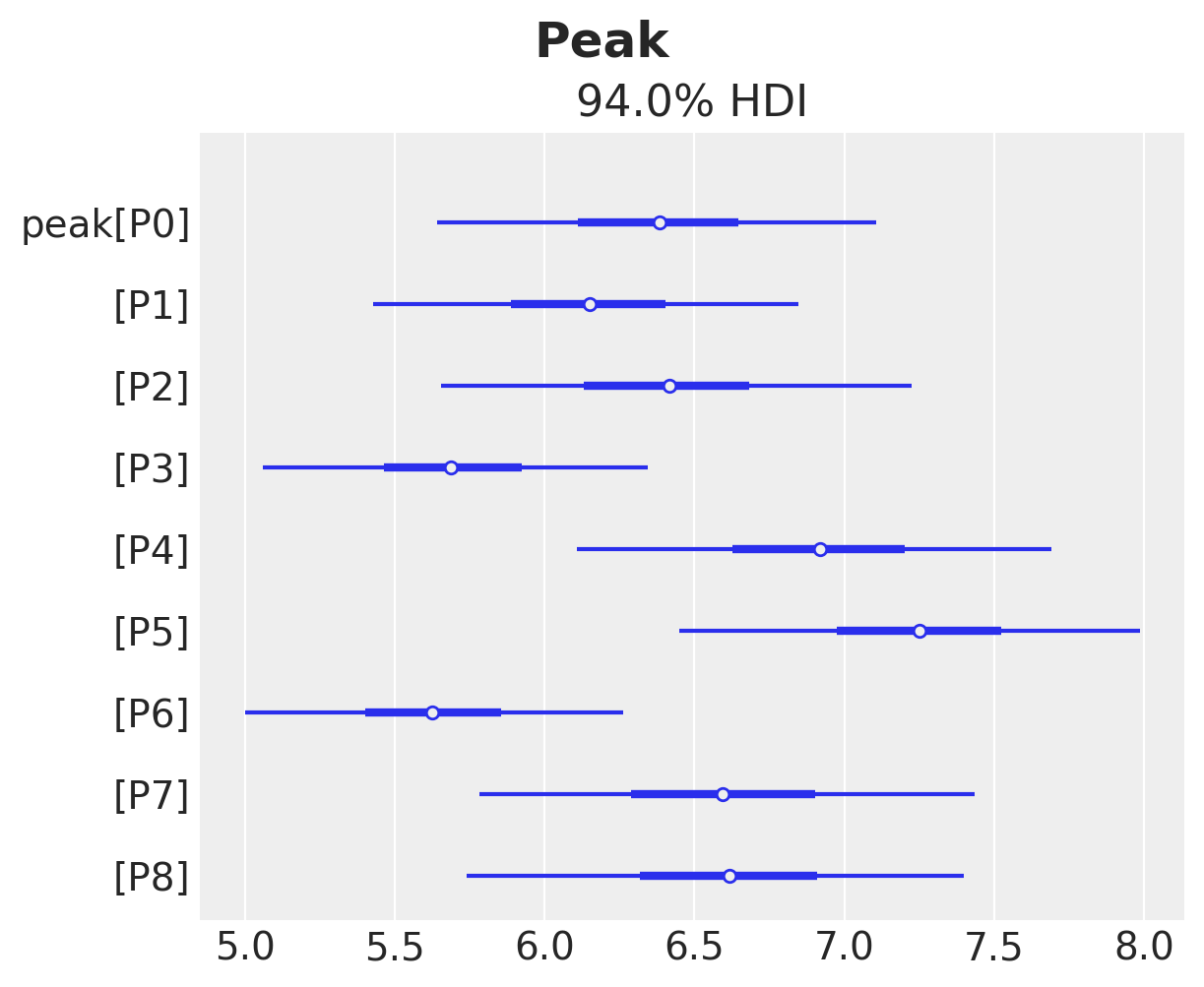
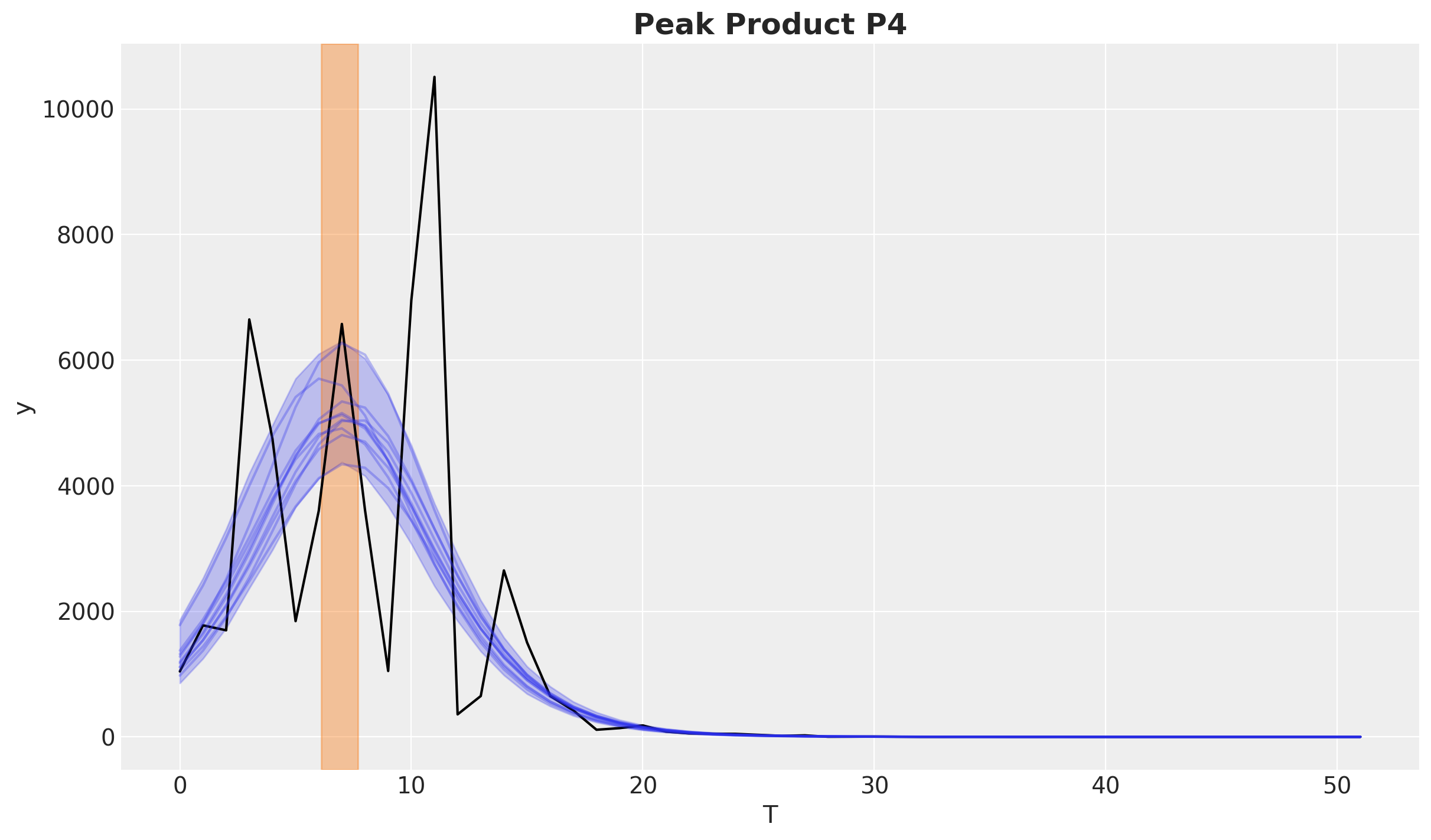
This fits the observed data quite well. Let’s see for example the product P4.
fig, ax = plt.subplots()
product_id = 4
bass_data[:, product_id].plot(ax=ax, color="black")
idata["posterior"]["adopters"].sel(product=f"P{product_id}").pipe(
plot_curve, {"T"}, axes=ax
)
peak_hdi = az.hdi(idata["posterior"]["peak"].sel(product=f"P{product_id}"))["peak"]
ax.axvspan(
peak_hdi.sel(hdi="lower").item(),
peak_hdi.sel(hdi="higher").item(),
color="C1",
alpha=0.4,
)
ax.set_title(f"Peak Product {products[product_id]}", fontsize=18, fontweight="bold");
%load_ext watermark
%watermark -n -u -v -iv -w -p nutpie,pymc_marketing,pytensor
Last updated: Wed May 21 2025
Python implementation: CPython
Python version : 3.10.16
IPython version : 8.34.0
nutpie : 0.14.3
pymc_marketing: 0.13.1
pytensor : 2.30.3
pymc_marketing: 0.13.1
xarray : 2025.3.1
arviz : 0.21.0
numpy : 1.26.4
pandas : 2.2.3
matplotlib : 3.10.1
pymc : 5.22.0
Watermark: 2.5.0
